by Emily Singer
March 24, 2016
from QuantaMagazine Website
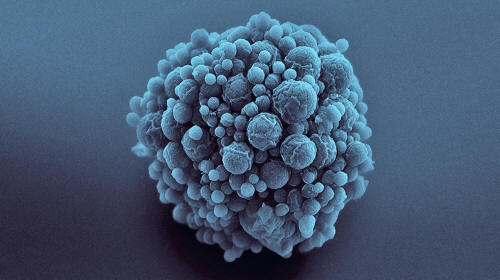
The ‘syn3.0’ cells
contain the minimum number of genes
needed for life.
Scientists have created a synthetic organism
that possesses only the genes it needs to survive.
But they have no idea
what roughly a third of those genes do.
Peel away the layers of a house – the plastered walls, the slate roof, the hardwood floors – and you’re left with a frame, the skeletal form that makes up the core of any structure.
Can we do the same with life? Can scientists pare down the layers of complexity to reveal the essence of life, the foundation on which biology is built?
That’s what Craig Venter and his collaborators have attempted to do in a new study (Design and Synthesis of a Minimal Bacterial Genome) published today in the journal Science. Venter’s team painstakingly whittled down the genome of Mycoplasma mycoides, a bacterium that lives in cattle, to reveal a bare-bones set of genetic instructions capable of making life.
The result is a tiny organism named ‘syn3.0‘ that contains just 473 genes (by comparison, E. coli has about 4,000 to 5,000 genes, and humans have roughly 20,000.)
Yet within those 473 genes lies a gaping hole. Scientists have little idea what roughly a third of them do.
Rather than illuminating the essential components of life, syn3.0 has revealed how much we have left to learn about the very basics of biology.
“To me, the most interesting thing is what it tells us about what we don’t know,” said
Jack Szostak, a biochemist at Harvard University who was not involved in the study.
“So many genes of unknown function seem to be essential.”
“We were totally surprised and shocked,” said Venter, a biologist who heads the J. Craig Venter Institute in La Jolla, Calif., and Rockville, Md., and is most famous for his role in mapping the human genome.
The researchers had expected some number of unknown genes in the mix, perhaps totaling five to 10 percent of the genome.
“But this is truly a stunning number,” he said.
The seed for Venter’s quest was planted in 1995, when his team deciphered the genome of Mycoplasma genitalium, a microbe that lives in the human urinary tract.
When Venter’s researchers started work on this new project, they chose M. genitalium – the second complete bacterial genome to be sequenced – expressly for its diminutive genome size. With 517 genes and 580,000 DNA letters, it has one of the smallest known genomes in a self-replicating organism.
(Some symbiotic microbes can survive with just 100-odd genes, but they rely on resources from their host to survive.)
M. genitalium‘s trim package of DNA raised the question: What is the smallest number of genes a cell could possess?
“We wanted to know the basic gene components of life,” Venter said. “It seemed like a great idea 20 years ago – we had no idea it would be a 20-year process to get here.”

JCVI
Clyde A. Hutchison,
a biologist at JCVI who led the new study,
has been researching mycoplasma bacteria as models
for the minimal cell since 1990.
Minimal Design
Venter and his collaborators originally set out to design a stripped-down genome based on what scientists knew about biology.
They would start with genes involved in the most critical processes of the cell, such as copying and translating DNA, and build from there. But before they could create this streamlined version of life, the researchers had to figure out how to design and build genomes from scratch.
Rather than editing DNA in a living organism, as most researchers did, they wanted to exert greater control – to plan their genome on a computer and then synthesize the DNA in test tubes.
In 2008, Venter and his collaborator Hamilton Smith created the first synthetic bacterial genome by building a modified version of M. genitalium‘s DNA.
Then in 2010 they made the first self-replicating synthetic organism, manufacturing a version of M. mycoides‘ genome and then transplanting it into a different Mycoplasma species.
The synthetic genome took over the cell, replacing the native operating system with a human-made version.
The synthetic M. mycoides genome was mostly identical to the natural version, save for a few genetic watermarks – researchers added their names and a few famous quotes, including a slightly garbled version of Richard Feynman‘s assertion,
“What I cannot create, I do not understand.”
With the right tools finally in hand, the researchers designed a set of genetic blueprints for their minimal cell and then tried to build them.
Yet “not one design worked,” Venter said.
He saw their repeated failures as a rebuke for their hubris.
Does modern science have sufficient knowledge of basic biological principles to build a cell?
“The answer was a resounding no,” he said.
So the team took a different and more labor-intensive tack, replacing the design approach with trial and error.
They disrupted M. mycoides‘ genes, determining which were essential for the bacteria to survive. They erased the extraneous genes to create ‘syn3.0’, which has a smaller genome than any independently replicating organism discovered on Earth to date.
What’s left after trimming the genetic fat?
The majority of the remaining genes are involved in one of three functions:
producing RNA and proteins, preserving the fidelity of genetic information, or creating the cell membrane. Genes for editing DNA were largely expendable.
But it is unclear what the remaining 149 genes do...
Scientists can broadly classify 70 of them based on the genes’ structure, but the researchers have little idea of what precise role the genes play in the cell.
The function of 79 genes is a complete mystery.
“We don’t know what they provide or why they are essential for life – maybe they are doing something more subtle, something obviously not appreciated yet in biology,” Venter said.
“It’s a very humbling set of experiments.”
***
Synthetic Biology
Venter envisions syn3.0 as a cellular chassis that scientists can build on.
Researchers can embellish the genome to create new organisms, which could help them to better understand stages of evolution lost to time.
“In theory, we should be able to add genes back to [syn3.0] to recapitulate key parts of evolution,” Venter said.
For example, they might try to create more advanced bacteria, or even to convert the basic chassis into different biological classes altogether.
“We could reduce billions of years of evolution to maybe years or months or weeks,” he said.
Venter and his collaborators also plan to use the cells for industrial purposes, designing cells that can produce pharmaceuticals or other chemicals.
“We have one cell in production to make omega-3s more efficiently than it can be isolated from fish,” Venter said.
One of the challenges in synthetic biology – the quest to engineer cells for specific purposes – has been that living organisms behave unpredictably.
Theoretically, a minimal cell would provide an engineering advantage because it has fewer unpredictable components. It’s not yet clear whether this will prove true.
Most efforts in synthetic biology employ existing microbes, such as
E. coli, and scientists may not yet see a good reason to switch.
Venter’s team is eager to figure out what the mystery genes do, but the challenge is multiplied by the fact that these genes don’t resemble any other known genes.
One way to investigate their function is to engineer versions of the cell in which each of these genes can be turned on and off.
When they’re off,
“what’s the first thing to get messed up?” Szostak said. “You can try to pin it to general class, like metabolism or DNA replication.”
Dwindling to Zero
Venter is careful to avoid calling ‘syn3.0’ a universal minimal cell.
If he had done the same set of experiments with a different microbe, he points out, he would have ended up with a different set of genes. In fact, there’s no single set of genes that all living things need in order to exist.
When scientists first began searching for such a thing 20 years ago, they hoped that simply comparing the genome sequences from a bunch of different species would reveal an essential core shared by all species.
But as the number of genome sequences blossomed, that essential core disappeared.
In 2010, David Ussery, a biologist at Oak Ridge National Laboratory in Tennessee, and his collaborators compared 1,000 genomes.
They found that not a single gene is shared across all of life.
“There are different ways to have a core set of instructions,” Szostak said.
Moreover, what’s essential in biology depends largely on an organism’s environment.
For example, imagine a microbe that lives in the presence of a toxin, such as an antibiotic. A gene that can break down the toxin would be essential for a microbe in that environment. But remove the toxin, and that gene is no longer essential.
Venter’s minimal cell is a product not just of its environment, but of the entirety of the history of life on Earth.
Sometime in biology’s 4-billion-year record, cells much simpler than this one must have existed.
“We didn’t go from nothing to a cell with 400 genes,” Szostak said.
He and others are trying to make more basic life-forms that are representative of these earlier stages of evolution.
Some scientists say that this type of bottom-up approach is necessary in order to truly understand life’s essence.
“If we are ever to understand even the simplest living organism, we have to be able to design and synthesize one from scratch,” said
Anthony Forster, a biologist at Uppsala University in Sweden.
“We are still far from this goal.”

by Tony Cartalucci
January 5, 2013
from LocalOrg Website
Professor Jamie Davies walks an audience through the coming synthetic biology revolution.
Comparing it to the personal computer revolution of the 70’s and 80’s, Professor Davies explains the lessons learned and how they can be applied to developing an open and constructive use of synthetic biology.
What is synthetic biology (below video)?
It is the next step in genetic engineering – not simply copying and pasting genetic code from one life form to another, but creating entirely new genetic sequences, and thus entirely new life forms.
Already, competitions like MIT’s iGEM, pit universities and even high schools against one another as they develop new forms of synthetic biology using “biobricks“-open-source, standardized components that can be interchanged in the designing of a synthetic life form just as engineers use standardized parts to construct machines and buildings today.
Examples of life forms created include bacteria that change color in the presence of toxins in the environment, and yeasts that are transformed into microscopic factories producing medicine.
The implications go further still – with the ability to read a human genome and understand, and as synthetic biology matures, it may be possible in the future to read one’s genetic code, correct the errors that accumulate over time, and create repaired code that is reintroduced into the body via gene therapy.
This would mitigate degenerative conditions stemming from aging, and all the diseases such deterioration invites, including cancers.
Professor Davies most important talking point however, centers on the DIYbio movement and how regular people are actively participating in this revolution – and how such participation is essential in keeping this technology free and available to all.
He points out local DIYbio groups springing up around the world and how amateurs and professionals alike are teaming up to advance this new field of study. He surmises that the great interest in synthetic biology is the ability to actually build things (life forms in this case).
While there are threats of people abusing this technology, just as is the case with any other form of technology – just like with information technology and computing, the more people that are involved and actively participating and the more decentralized the infrastructure is, the greater our ability collectively is at defending against inevitable abuses.
The greatest danger is if this technology remains in the hands of large institutions, corporations, and tangled up in a web of contrived “intellectual property” claims.
Ultimately, one walks away from Professor Davies’ talk with a sense of optimism, but also with a call for action.
If we are to harness the full potential of this technology, we will have to roll up our sleeves and get involved. If we fail to do this, the technology will be patented, black-boxed, copyrighted, and monopolized.
The fear and real dangers produced by genetic engineering today, stems from the fact that immensely corrupt, centralized corporations monopolize the technology and willfully and consistently abuse it to expand profits and control over the very substance of life.
The emerging field of DIYbio and synthetic biology gives us a chance to level the playing field and put both the technology and its benefits where they belong – in the people’s hands.
Prof. Jamie Davies
Synthetic Biology – The Potential and The problems of Re-Engineering Life